2D RCM-1 Finite Rate Tutorial
This tutorial covers the basics of setting up and running the RCM-1 combustion case, which is commonly used in the CFD community for testing combustion models for turbulence/chemistry interaction. In this example, we employ a 2-D, one degree pie-slice grid and use RANS turbulence modeling to obtain a steady-state solution to the problem using finite-rate chemistry. This RANS solution is then used as an initial condition for an unsteady detached-eddy simulation (DES). The files that are needed to run this case can be extracted from a gzipped archive located here. The files will extract into a directory structure that mirrors this case’s location in the tutorials repository.
Geometry and Boundary Conditions
The geometry and boundary conditions for this case are shown below. Fuel, composed of 40.18% H2 and 59.82% H2O by mass, enters the domain at the upper left inlet. Oxidizer, composed of 94.49% O2 and 5.51% H2O by mass, enters the domain at the lower left inlet. With the exception of the upper combustion chamber wall which has a specified temperature profile, all walls are specified as adiabatic boundaries. Flow at the nozzle exit becomes supersonic before steady state is achieved, and is specified as an extrapolated-pressure outlet for the duration of the simulation.

Geometry and boundary conditions for the RCM-1 finite-rate tutorial.
Grid
A mixed triangular/quadrilateral cell grid was first generated using SolidMesh. This was then extruded by SolidMesh into a one degree pie-slice grid comprising 204,106 prismatic/hexahedral cells. Particular emphasis during the grid-generation phase was place on proper resolution of the wall boundary layers, the entrance region to the combustion chamber and the combustion post region where fuel and oxidizer first meet and the nozzle throat region. Close-up images of select regions of the grid are shown below.

The upstream region of the RCM-1 grid.
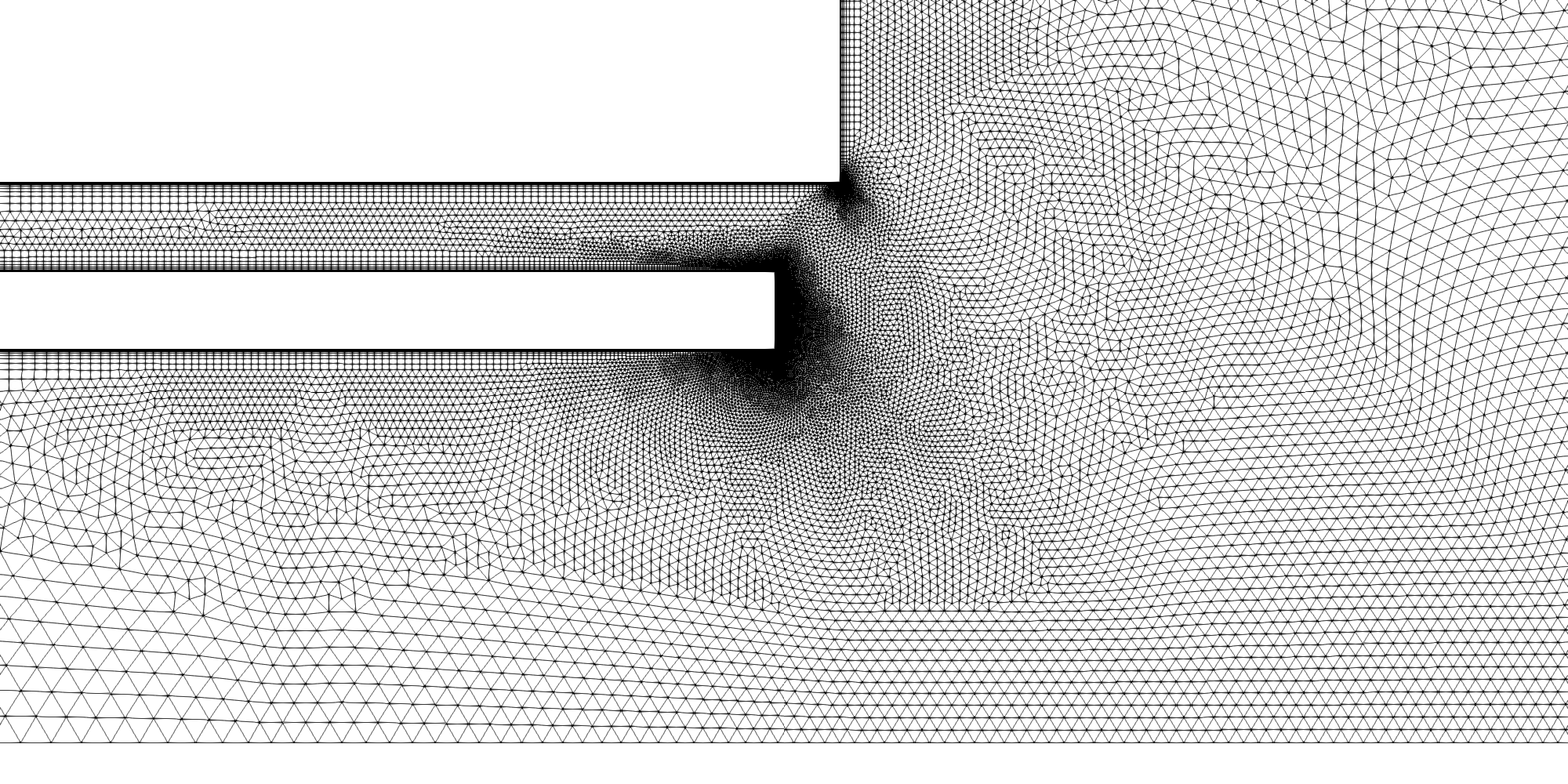
The combustion post region of the RCM-1 grid.
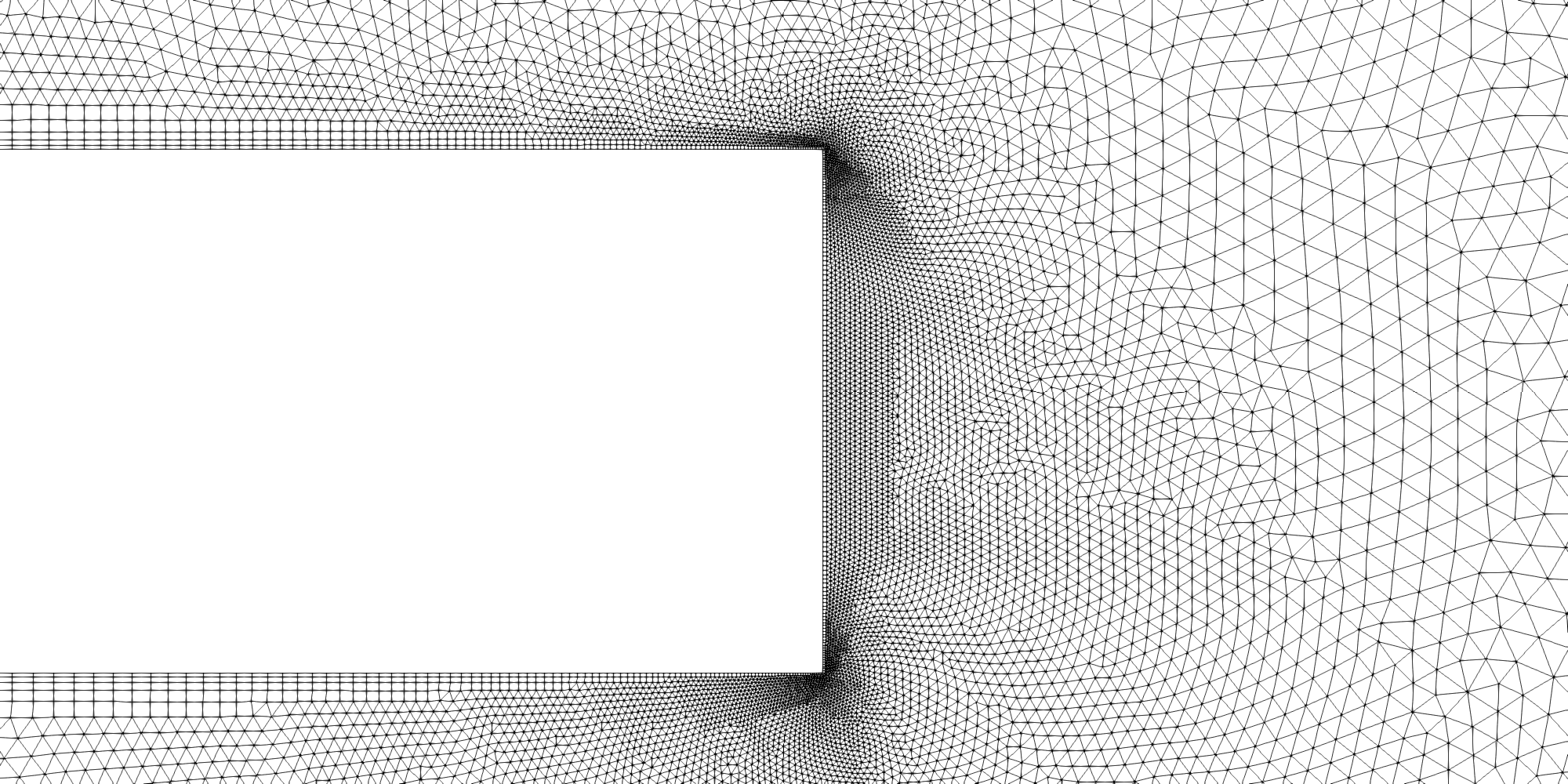
Zoomed view of the combustion post region of the RCM-1 grid.
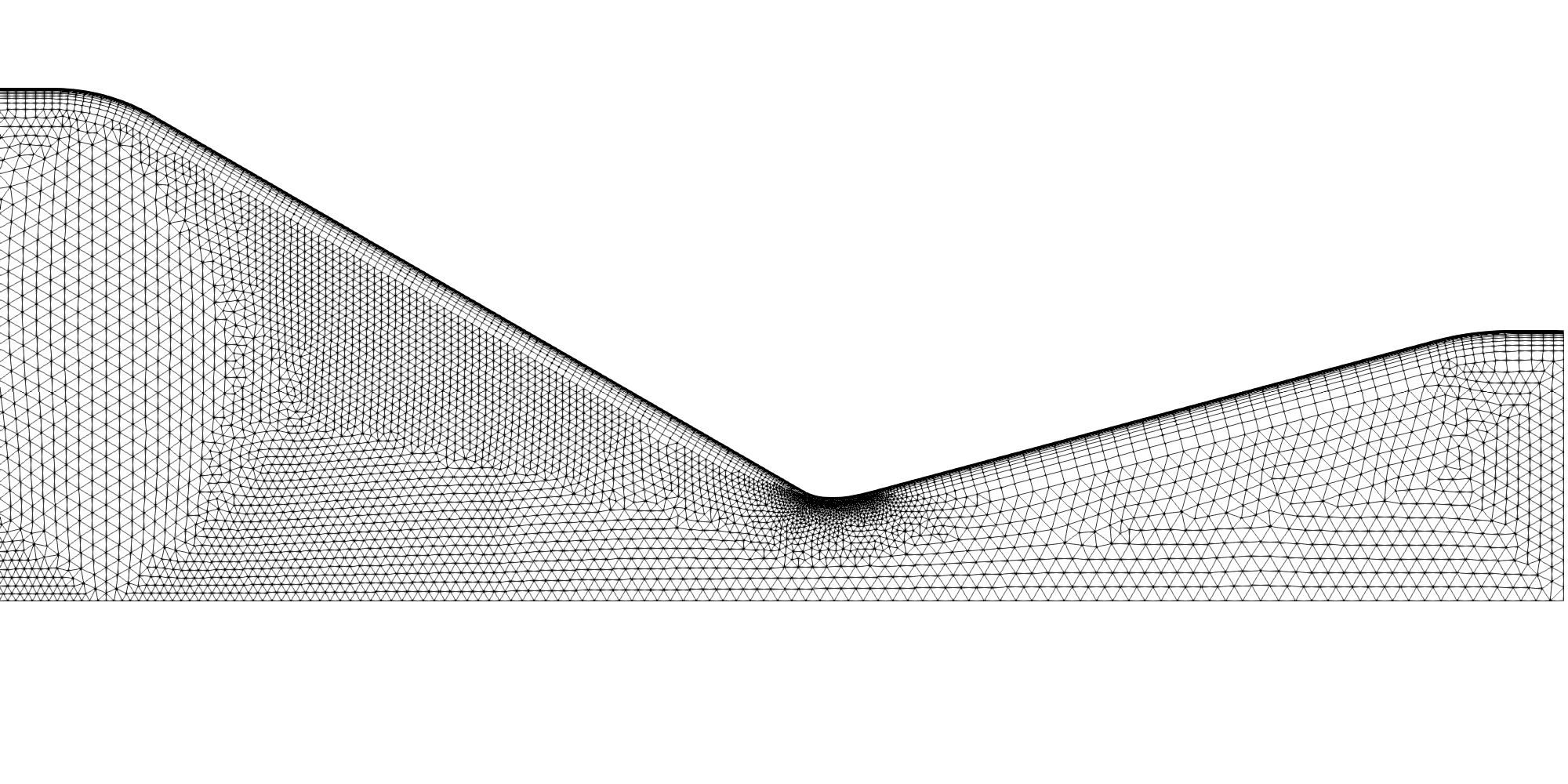
The nozzle region of the RCM-1 grid.
Run Control File Setup for RANS Simulation
The run control file for the RANS simulation is shown below.
{
// Grid file information
grid_file_info: <pieSlice, file_type=VOG>
// Boundary conditions
boundary_conditions:
<
Oxidizer_Inlet=subsonicInlet(mdot=2.51243E-04 kg/s, T=700.0 K, mixture=[O2=0.9449,H2O=0.0551], k=4.2, omega=12900.0),
Fuel_Inlet=subsonicInlet(mdot=9.1978e-05 kg/s, T=811.0 K, mixture=[H2=0.4018,H2O=0.5982], k=0.5, omega=37950.0),
Outlet=extrapolatedPressureOutlet,
Face_Plate=noslip(adiabatic),
Post_Tip=noslip(adiabatic),
Oxidizer_Walls=noslip(adiabatic),
Fuel_Walls=noslip(adiabatic),
ChamberWalls=noslip(T=cartesian(file="bc_temp.dat")),
Nozzle=noslip(adiabatic),
Back_PieSlice=symmetry,
Front_PieSlice=symmetry
>
// Initial condition
initialCondition: <v=[0.0 m/s,0.0 m/s,0.0 m/s], p=5.4e+06 Pa, T=1500.0 K, mixture=[H2O=1.0], k=0.5, omega=37950.0>
// Flow properties
flowRegime: turbulent
flowCompressibility: compressible
// Equation of state and transport properties
chemistry_model: H2
thermodynamic_model: curve_fit
transport_model: chemkin
diffusion_model: unityLewisNumber
// Time-stepping
timeIntegrator: BDF
timeStep: 1.0e-05
numTimeSteps: 20001
convergenceTolerance: 1.0e-19
maxIterationsPerTimeStep: 11
// Inviscid flux
inviscidFlux: FOU
turbulenceInviscidFlux: FOU
// Gradient limiting
limiter: venkatakrishnan
// Linear solver options
linearSolverTolerance: 1.0e-02
hypreSolverName: AMG
hypreStrongThreshold: 0.9
// Momentum equation
momentumEquationOptions: <linearSolver=SGS,relaxationFactor=0.5,maxIterations=5>
// Pressure equation
pressureCorrectionEquationOptions: <linearSolver=HYPRE,relaxationFactor=0.3,maxIterations=20>
pressureBasedMethod: SIMPLEC
// Energy equation
energyEquationOptions: <linearSolver=SGS,relaxationFactor=0.5,maxIterations=5,form=totalEnergy>
TclipMin: 500.0
TclipMax: 5000.0
// Turbulence equations
turbulenceEquationOptions: <model=menterSST2003,linearSolver=SGS,relaxationFactor=0.5,maxIterations=5>
turbulentPrandtlNumber: 0.5
turbulentSchmidtNumber: 0.5
kFreestream: 4.2
omegaFreestream: 12900
eddyViscosityLimit: 10000.0
// Species equations
speciesEquationOptions: <linearSolver=SGS,relaxationFactor=0.5,maxIterations=5>
operatorSplitting: strang
operatorSplittingEOS: 2
cvodeAbsoluteTolerance: 1.0e-10
cvodeRelativeTolerance: 1.0e-04
speciesRenormalization: off
speciesFaceDiffusion: conservative
speciesFaceReconstruction: conservative
Tf: 300.0
// Printing, plotting, and restart parameters
print_freq: 1000
plot_freq: 1000
plot_modulo: 0
plot_output: e, viscosityRatio, pResidual, eResidual
boundary_plot_freq: 1000
restart_freq: 1000
restart_modulo: 0
}
In the following, we briefly discuss some of the key choices made in setting the run control file parameters for this case.
In the
grid_file_infovariable, we have placed thepieSliceoption, since we are solving a 2-D flow on a pie-slice grid. This option causes Stream to zero out the velocity field in the z-coordinate direction for all computations. When using this option, it is important that the grid be oriented in the z=0 plane with the center of the pie-slice at z=0.The temperature profile for the
ChamberWallsboundary condition segment has been set using thecartesianspecification detailed in the Stream users’ guide. For this case, the filebc_temp.datis set up to specify a temperature versus x-coordinate profile because the chamber top wall coordinates vary along the x-coordinate direction.The simulation time step value of 5.0e-06 was chosen by dividing the flow-through time (from oxygen inlet to nozzle exit) of the flow along the axis (estimated to be approximatedly 5.0e-3 seconds) by 1000 time steps. Thus the
numTimeStepsvalue of 20,000 results in approximately 20 flow-throughs, which is sufficient for all unsteady flow behaviour to dampen out.Since we are interested in the steady-state answer, we can set all fluxes (
inviscidFluxandturbulenceInviscidFlux) toFOUand run the simulation to steady-state. This will give us a baseline spatially low-order solution that we can then restart from with higher order spatial flux schemes.Relaxation factors for the governing equations have been set to relatively low values to overcome the poor initial condition. The turbulence equations relaxation factor has been kept relatively high in order to quickly develop a turbulent viscosity which can stabilize the solution convergence.
Running the RANS Simulation
In the tutorial main directory are three files that are required to run the RANS and DES cases.
case.vog: the grid
bc_temp.dat: a temperature profile along the upper wall of the combustor
H2.mdl: the chemistry model file that contains species thermodynamic and reaction data
H2.tran: a transport property file containing the species molecular weights, viscosities, thermal conductivities, and diffusion coefficients
Also in this directory is a sub-directory called rans/ , which contains the run control file for this case.
To run the RANS case, copy the case.vog, bc_temp.dat, H2.mdl, and H2.tran files to the rans/ directory.
For this case, the grid has around 206,000 cells. Stream has parallel efficiency down to 5,000 cells per processor, so you will
want to run using less than 40 processes for the value passed to the -np argument in the mpirun command shown below. Using fewer
processes is fine and will only result in an increased time for the solution to be generated. Enter the rans/ directory and execute
the code using the following command.
mpirun -np 40 <path_to_stream_exec> --scheduleoutput -q solution case >& run.log_0 &
The RANS case will run until the number of timesteps specified in the case.vars file is reached. Restart checkpoint solutions will be
written to the restart/ directory at the number of timesteps specified in the case.vars file for the restart_freq variable.
Once the RANS case is completed, the RANS solution can be used as an initial condition to the DES case. The details about restarting a DES
case from a RANS solution are discussed in the DES section that follows the RANS section.
RANS Results
The extract utility is used to view the results of a simulation. For this case we view the simulation at the 10,000th time step. In the
run directory, use the following command: extract -vtk case 10000 r v P t m fH2O fOH . This will generate a solution directory with files
in the Paraview VTK format which can be opened by Paraview. The case name is case and the time step is 10000, and the variables to extract
are the density(r), velocity(v), pressure(P), temperature(t), mach number(m), H2O mass fraction(fH2O), and OH mass fraction(fOH).
Shown below are select plots of the solution variables.
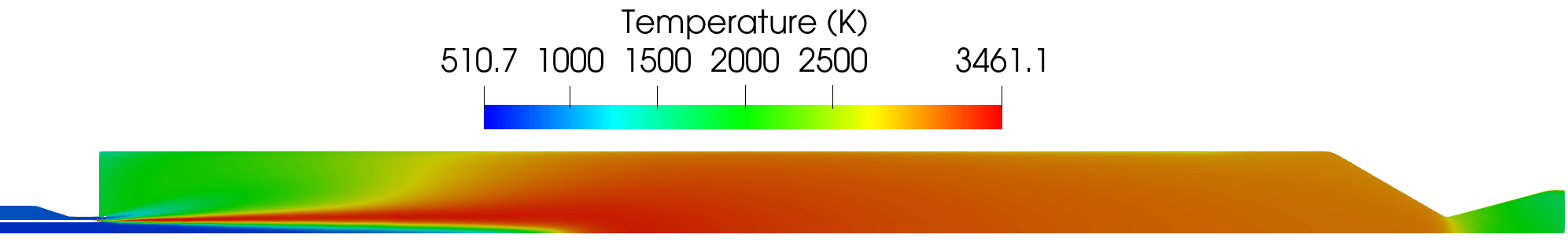
Contour plot of the temperature field for the RANS simulation.
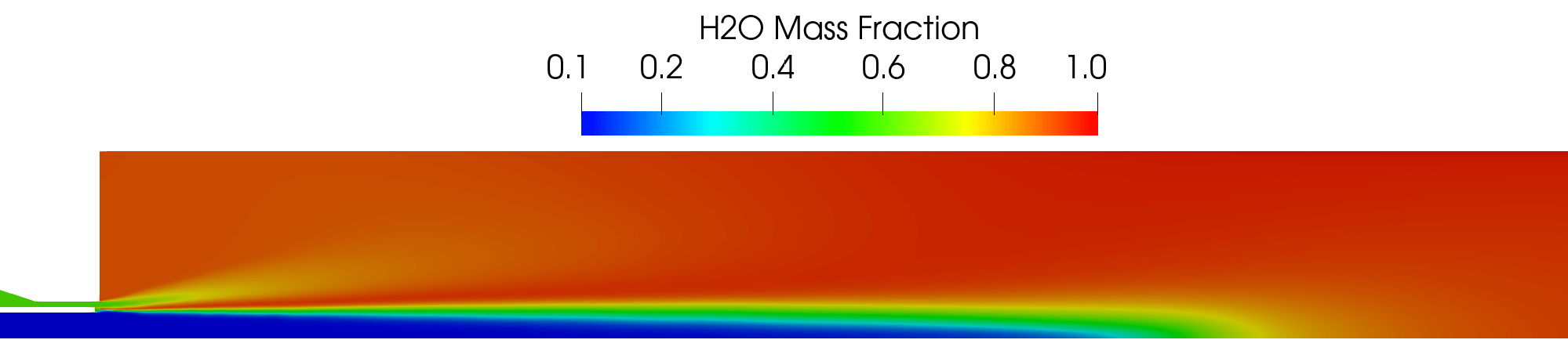
Contour plot of the H2O mass fraction field for the RANS simulation.
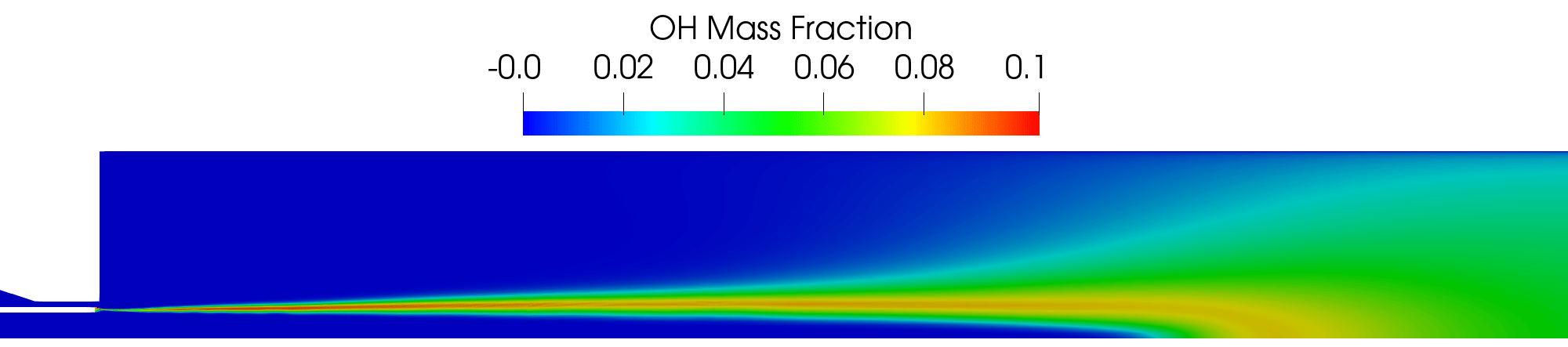
Contour plot of the OH mass fraction field for the RANS simulation.
Run Control File Setup for DES Simulation
The run control file for the DES simulation is shown below.
{
// Grid file information
grid_file_info: <pieSlice, file_type=VOG>
// Boundary conditions
boundary_conditions:
<
Oxidizer_Inlet=subsonicInlet(mdot=2.51243E-04 kg/s, T=700.0 K, mixture=[O2=0.9449,H2O=0.0551], k=4.2, omega=12900.0),
Fuel_Inlet=subsonicInlet(mdot=9.1978e-05 kg/s, T=811.0 K, mixture=[H2=0.4018,H2O=0.5982], k=0.5, omega=37950.0),
Outlet=extrapolatedPressureOutlet,
Face_Plate=noslip(adiabatic),
Post_Tip=noslip(adiabatic),
Oxidizer_Walls=noslip(adiabatic),
Fuel_Walls=noslip(adiabatic),
ChamberWalls=noslip(T=cartesian(file="bc_temp.dat")),
Nozzle=noslip(adiabatic),
Back_PieSlice=symmetry,
Front_PieSlice=symmetry
>
// Initial condition
initialCondition: <v=[0.0 m/s,0.0 m/s,0.0 m/s], p=5.4e+06 Pa, T=1500.0 K, mixture=[H2O=1.0], k=0.5, omega=37950.0>
// Flow properties
flowRegime: turbulent
flowCompressibility: compressible
// Equation of state and transport properties
chemistry_model: H2
thermodynamic_model: curve_fit
transport_model: chemkin
diffusion_model: unityLewisNumber
// Time-stepping
timeIntegrator: CN
CNBDFBlend: 0.9
timeStep: 3.0e-07
numTimeSteps: 190001
convergenceTolerance: 1.0e-19
maxIterationsPerTimeStep: 21
// Inviscid flux
inviscidFlux: SLAU
slau_mhat: 2
slau_reconstruct: 2
slau_pfactor: 0.0
turbulenceInviscidFlux: SOU
// Gradient limiting
limiter: venkatakrishnan
// Linear solver options
linearSolverTolerance: 1.0e-02
hypreSolverName: AMG
hypreStrongThreshold: 0.9
// Momentum equation
momentumEquationOptions: <linearSolver=SGS,relaxationFactor=0.7,maxIterations=5>
// Pressure equation
pressureCorrectionEquationOptions: <linearSolver=HYPRE,relaxationFactor=0.5,maxIterations=20>
pressureBasedMethod: SIMPLEC
// Energy equation
energyEquationOptions: <linearSolver=SGS,relaxationFactor=0.7,maxIterations=5,form=totalEnergy>
TclipMin: 500.0
TclipMax: 5000.0
// Turbulence equations
turbulenceEquationOptions: <model=menterSST2003,des=IDDES2012,linearSolver=SGS,relaxationFactor=0.7,maxIterations=5>
turbulentPrandtlNumber: 0.5
turbulentSchmidtNumber: 0.5
kFreestream: 4.2
omegaFreestream: 12900
eddyViscosityLimit: 10000.0
// Species equations
speciesEquationOptions: <linearSolver=SGS,relaxationFactor=0.7,maxIterations=5>
operatorSplitting: strang
operatorSplittingEOS: 2
cvodeAbsoluteTolerance: 1.0e-10
cvodeRelativeTolerance: 1.0e-04
speciesRenormalization: off
speciesFaceDiffusion: conservative
speciesFaceReconstruction: conservative
Tf: 300.0
// Printing, plotting, and restart parameters
print_freq: 1000
plot_freq: 1000
plot_modulo: 0
plot_output: e, viscosityRatio
boundary_plot_freq: 1000
restart_freq: 5000
restart_modulo: 0
}
In the following, we briefly discuss some of the key choices made in setting the run control file parameters for the DES case.
While this case is being restarted from a RANS solution, note that we must still specify an initial condition in the run control file to circumvent Stream’s run control file consistency checking coding from returning an error. Upon restart, however, the values specified in the variable
initialConditionwill be ignored.The DES simulation is a time-accurate simulation. As such, we have selected the blended Crank-Nicolson/BDF scheme with a blending factor of 0.9, which in our experience achieves time accuracy at a slighly larger time step than the BDF2 time-differencing scheme. Note that a relatively small time step of 2.0e-07 is still required for stability and time-accuracy purposes compared to the value chosen for the RANS simulation. Here we have used 21 iterations per time step, however, based on evaluation of the residual convergence in the run log file, one could probably use less and save CPU time.
For the inviscid fluxes, we have chosen the SLAU scheme, which is generally superior to SOU for compressible flows, and have also used second-order inviscid flux reconstruction for the turbulence and flamelet equations, in order to avoid the excess spatial dissipation one would get using the FOU scheme.
For the pressure-correction equation, one should always use the HYPRE linear solver. The parameters in the linear solver section apply only to the HYPRE solver and are not used by the SGS linear solver which is being employed here for the momentum, total energy and flamelet equations. Here we select the Algebraic Multi-Grid (AMG) solver, which is superior in general to the GMRES solver in HYPRE. We set the linear solver tolerance to a relatively high value of 1.0e-02 to avoid over solving the linear system for the pressure-correction equation. The
hypreStrongThresholdparameter is set to a relatively high value (range 0-1,default is 0.5) to further minimize the work per linear solver iteration.At the end of the run control file, for tutorial purposes, we show several variables (
hypreSolverOutput,massTurnOverTimeOutput,residualMaximumOutput) that can be used to monitor the health of the solution process. See the Stream users’ guide for details on these variables. For lengthy simulations (hundreds of thousands of timesteps) one should ensure that the values for these three diagnostic variables areoff, in order to prevent debug files in the/debugdirectory from becoming excessively large (gigabytes).
Running the DES Simulation
In the tutorial main directory are the following files that are required to run the DES cases.
case.vog: the grid
bc_temp.dat: a temperature profile along the upper wall of the combustor
H2.mdl: the chemistry model file that contains species thermodynamic and reaction data
H2.tran: a transport property file containing the species molecular weights, viscosities, thermal conductivities, and diffusion coefficients
Also in this directory is a sub-directory called des/ , which contains the run control file for this case.
To run the DES case, first copy the case.vog, bc_temp.dat, H2.mdl, and H2.tran files to the des/ directory. In addition,
you will create a new directory in the des/ directory called restart/. This directory is where you will place a set of
RANS restart files for the DES simulation to restart from. For this case, the RANS simulation was run to 10,000 timesteps and a set of
restart files were written at the 10,000th timestep. Go to the rans/ directory and perform a copy like the one shown below.
cp restart/*10000* ../des/restart/
For this case, the grid has around 206,000 cells. Stream has parallel efficiency down to 5,000 cells per processor, so you will
want to run using less than 40 processes for the value passed to the -np argument in the mpirun command shown below. Using fewer
processes is fine and will only result in an increased time for the solution to be generated. Enter the des/ directory and execute
the code using the following command.
mpirun -np 40 <path_to_stream_exec> --scheduleoutput -q solution case 10000 >& run.log_10000 &
The DES case will run an additional number of timesteps from the restart as specified by the numTimeSteps variable in the case.vars file. The case will
thus terminate at timestep 200,000. Restart checkpoint solutions will be written to the restart/ directory at the number of timesteps specified in
the case.vars file for the restart_freq``variable. The addition of the integer argument ``10000 in the command above tells Stream to use the solution
in the restart/ directory as the initial condition. In the command above, the file that the log information is redirected to is named run.log_10000. This
suffix is used to differentiate the log file from other log files that are generated when a user is restarting cases. The suffix notation prevents log file
information from previous simulations from being overwritten. The naming convention is not required, but is a good practice to follow when using Stream.
DES Results
The extract utility is used to view the results of a simulation. For this case we view the simulation at the 200,000th time step. In the
run directory, use the following command: extract -vtk case 200000 r v P t m fH2O fOH . This will generate a solution directory with files
in the Paraview VTK format which can be opened by Paraview. The case name is case and the time step is 200000, and the variables to extract
are the density(r), velocity(v), pressure(P), temperature(t), mach number(m), H2O mass fraction(fH2O), and OH mass fraction(fOH).
Shown below are select plots of the solution variables.

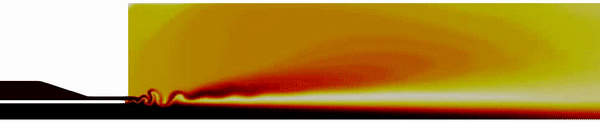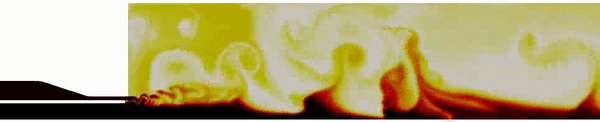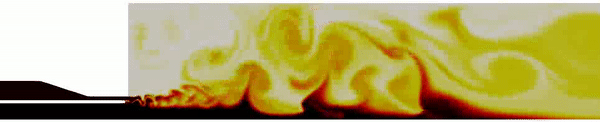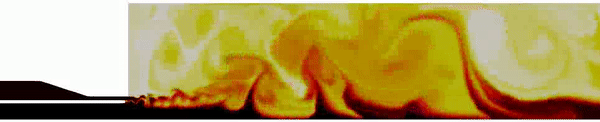
Contour plot of the temperature field for the DES simulation.
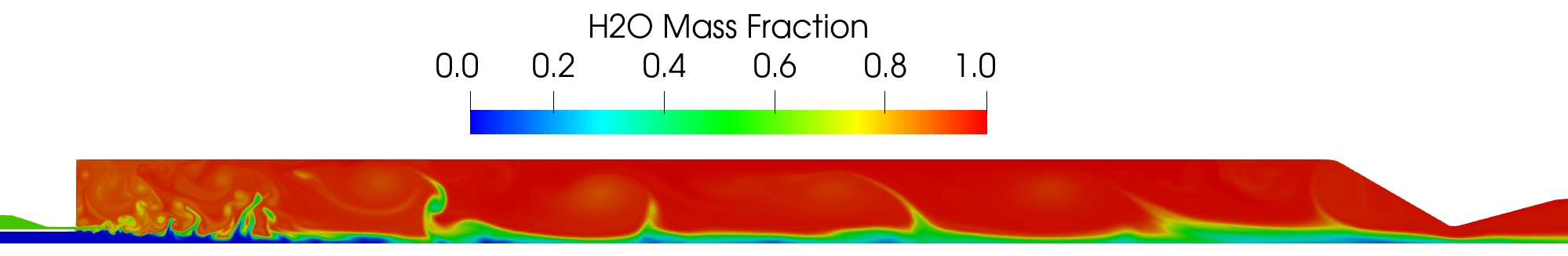
Contour plot of the H2O mass fraction field for the DES simulation.

Contour plot of the temperature field in the region near the injector for the DES simulation .
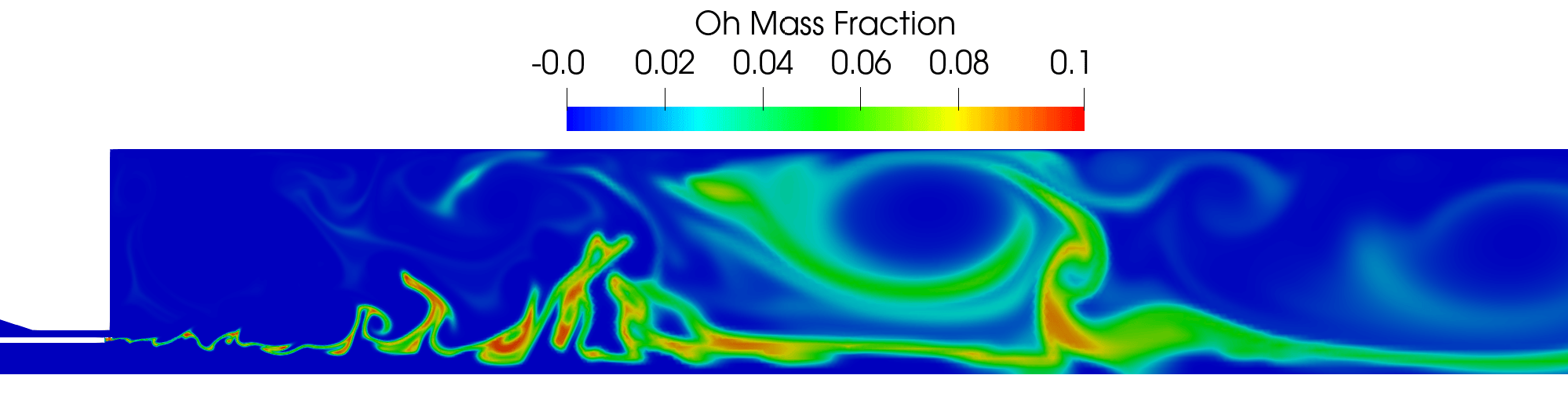
Contour plot of the OH mass fraction field in the region near the injector for the DES simulation .